PNGase F PRIME™ - Especially Optimized for Mass Spec Imaging
N-Zyme Products
N-Zyme Scientifics has been providing the research community with the best quality enzymes on the market since 2013. N-Zyme Scientifics has specifically engineered all of its enzymes for use in MALDI Mass Spectrometry Spatial Glycan Tissue Imaging. PNGase F PRIME™ has demonstrated superior efficacy in identifying glycan based biomarkers associated with disease progression in clinical tissue biopsy samples. Learn more about our most popular enzymes and their functionality in greater detail below. All of our products have been extensively formulated, optimized and utilized for mass spectrometry imaging-based experiments. Check back regularly for updates on other innovative new product offerings.
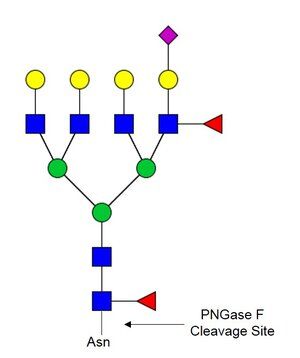
PNGase F PRIME™ GLYCOSIDASE (Liquid Format)
This enzyme is a proprietary mutant recombinant PNGase F, cloned from Flavobacterium meningosepticum and expressed and purified from E. coli. Excellent for use in high-end use applications especially those using or requiring UPLC/HPLC, Hydrophilic Interaction Chromatography (HILIC), and/or MALDI-Glycan Mass Spec Imaging. 1 Unit of enzyme will completely deglycosylate 10 µg of denatured RNase B following incubation for 10 minutes at 37°C. Impact on protein can be visualized by SDS-PAGE. Released glycans can be examined following labeling with the Waters RapiFluor-MS dye and analyzed by normal phase hydrophilic interaction chromatography (HILIC) using various HPLC/UPLC-based systems. Imaging of released glycans directly on tissue can be done following spraying of enzyme on tissue and incubation at 37°C for 1 hour. (At our standard concentration of 2mg/mL, one 50 µL vial can make 4 slides.) Glycans can be detected using instruments such as the Bruker Daltonics SolariX™ 7T Hybrid FTMS System, a Bruker Daltonics RapifleXTM MALDI Tissuetyper, or a Bruker Daltonics UltrafleXtreme MALDI-TOF/TOF mass spectrometer.
PNGase F PRIME-LY™ GLYCOSIDASE (Lyophilized Format)
PNGase F PRIME-LY™ Glycosidase is the same enzyme as our standard liquid format PNGase F PRIME™ but now available in a lyophilized form, which is perfectly suitable for use in solution-based analyses. Performance characteristics and applications in which it can be used remain exactly the same as with our standard liquid format PNGase F PRIME™ enzyme. This enzyme specifically removes N-linked oligosaccharides from glycoproteins.
PNGase F PRIME-LY™ ULTRA GLYCOSIDASE (Lyophilized Format)
PNGase F PRIME-LY™ ULTRA Glycosidase is optimized and specifically formulated for high resolution imaging in small structures or cells. This enzyme specifically removes N-linked oligosaccharides from glycoproteins. This is a high-performance enzyme that has been shown to require as few as 3,000 cells per replicate with 3-20% coefficient of variation to capture label-free N-glycan profiles (as shown in Angel et. al., J Proteome Res. 2019).
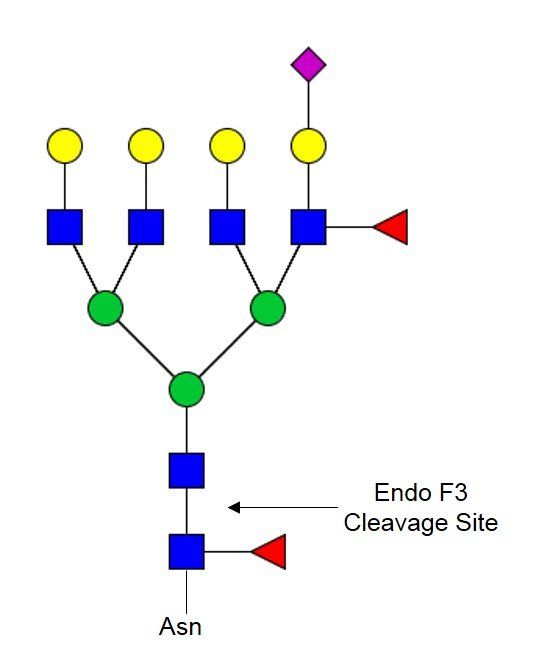
Endo F3 PRIME-LY™ (Lyophilized Format)
Endo-β-N-acetylglucosaminidase F3 (Endo F3) cleaves in β(1-4) link in between the two core GlcNAcs of asparagine linked glycans. Endo F3 cleaves this link on core-fucosylated structures. Endo F3 can be applied to workflows alone or in conjunction with PNGase F PRIME™ to allow for structural characterization of core-fucosylated glycans in tissues while maintaining spatial localization. Endo F3 PRIME-LY™ comes in a lyophilized format that is perfectly suitable for use in solution-based analyses.
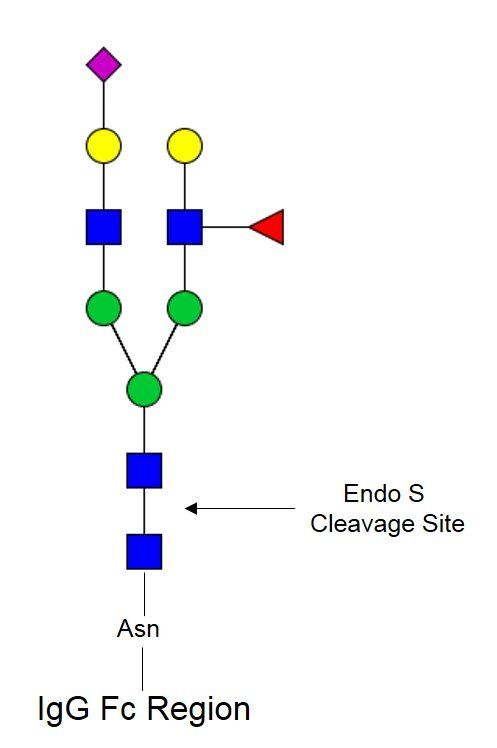
Endo S PRIME-LY™ (Lyophilized Format)
Endo-β-N-acetylglucosaminidase S (Endo S) cleaves the hydrolysis of the β(1-4) linkage between the two core GlcNAcs of asparagine linked biantennary complex-type glycans of IgG Fc regions. This endoglycosidase only deglycosylates IgG glycoforms and plays a central role in glycoengineering strategies for the development of IgG antibodies with improved therapeutic efficacy. Endo S PRIME-LY™ comes in a lyophilized format that is perfectly suitable for use in solution-based analyses.
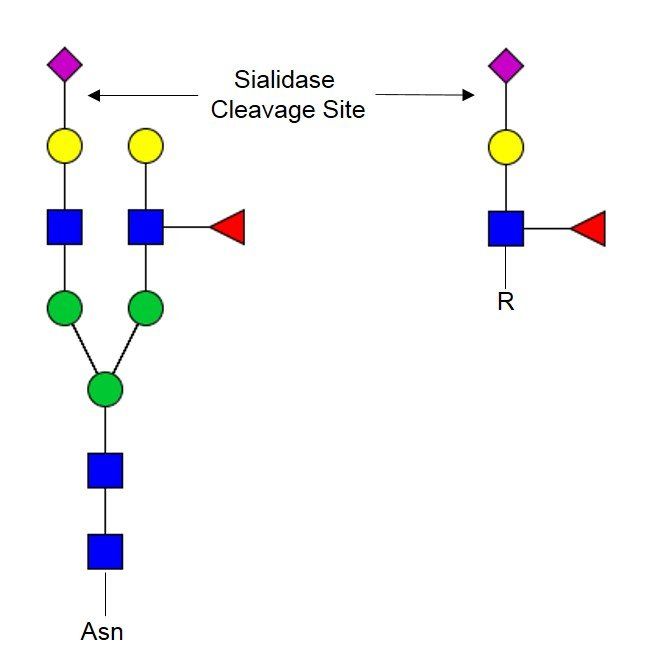
Sialidase PRIME-LY™ (Lyophilized Format)
Sialidases are a family of exoglycosidases that catalyze the cleavage of non-reducing sialic acid residues of mono- or oligosaccharide chains on glycoconjugates. Sialidase PRIME-LY™ comes in a lyophilized form, which is perfectly suitable for solution-based analyses. This enzyme cleaves α2,3-, α2,6- and α2,8- linked sialic acids. Because of its broad substrate specificity, Sialidase PRIME-LY™ is capable of completely removing sialic acids from glycoconjugates of a wide variety of biological materials (cells, antibodies, serum, tissues etc.).
For all your Glycan Tissue Imaging needs, there is GlycoPath's "Basic N-Glycan Imaging Kit". This Kit includes with its purchase, all protocols and data analysis information needed to perform glycan tissue analysis as done by the leaders and innovators in the field.
-
Basic N-Glycan Imaging Kit - Small (4 slides)
More InfoOur Basic N-Glycan Imaging Kit comes with our mass spectrometry imaging certified PNGaseF enzyme, PNGase F Activation Solution, CHCA Matrix, Positive Control Protein, and full detailed protocol. This kit has been validated to image glycans on up to 4 FFPE Tissue slides. While a protocol is included for Fresh Frozen tissue, the kit is currently only validated for use with FFPE tissues.
-
Basic N-Glycan Imaging Kit - Large (20 slides)
More InfoOur Basic N-Glycan Imaging Kit comes with our mass spectrometry imaging certified PNGaseF enzyme, PNGase F Activation Solution, CHCA Matrix, Positive Control Protein, and full detailed protocol. This kit has been validated to image glycans on up to 20 FFPE Tissue slides. While a protocol is included for Fresh Frozen tissue, the kit is currently only validated for use with FFPE tissues.